Author: Gregor Mathes
“[…] it’s impossible to divorce pure science from technology. They feed and stimulate each other.”
–Douglas Adams
When I was growing up, I imagined science offices as rooms filled with large bookshelves and occasionally a candle or two. The imagined picture of large amounts of paper combined with open fire was absolutely unsettling for me. Fortunately, reality turned out to be a bit different (even though I have seen some offices that indeed look like this, and with no fire extinguisher nearby as well).
When I was growing up, I imagined science offices as rooms filled with large bookshelves and occasionally a candle or two. The imagined picture of large amounts of paper combined with open fire was absolutely unsettling for me. Fortunately, reality turned out to be a bit different (even though I have seen some offices that indeed look like this, and with no fire extinguisher nearby as well).
Nowadays, the central object in a scientist’s office is the computer. We use computers to analyze data, visualize results, collaborate with co-workers all over the world, produce final manuscripts, and for many many more tasks. In contrast to a normal office job, where you set up the software configurations once and stick to it for years, scientists are confronted with an ever-changing software universe. And even if you have managed to find a specific setting of software tools that fit your scientific workflow; academic jobs are very short-lived and proprietary software might not be covered at your next position.

But a vast choice of software as well as pay-walls might not be the only issue a conservation palaeobiologist faces. Specific demands to software tools include (among others):
– well-established programming to guarantee robust and reproducible results
– ease of use for interdisciplinary research
– appealing output to communicate results with stake-holders
Throughout my scientific endeavors, I have worked my way through various software tools. Here I present you 10 open source software tools that I use daily. Each of these constitutes one module in a data-driven and project-oriented workflow and covers the needs mentioned above. Combined, they provide a foundation for open, accessible, and reproducible science. This includes that each tool is freely available and runs on all common operating systems (Windows, MacOS, and Linux distributions).
The Setup
The most important part in a project-oriented workflow begins before you have downloaded any software. Place all files in one folder. This will allow any software to directly access all files without changing the working directory, and will save you a lot of time. Now create subfolders (e.g., for code, manuscripts, figures etc.). By laying out the folder structure, you are forced to deeply think about the project itself. It is basically a sketch of your project before you start working on it. By having such a clean folder structure, the project can be moved around on your computer or onto other computers/laptops and will still work fine. It is the basic step towards a robust, reliable, and reproducible workflow.
The next step is to use a version control system. This will allow you to track changes and move “back in time” if necessary. Even if you delete all your data and code, you will still be able to access it using a version control system. Or if your analysis worked fine yesterday, but now gives you an error, you can go back to when everything was working. A free and open source distributed version control system widely used is Git. To incorporate Git in your workflow, you will need a hosting service like GitHub or GitLab. Since GitHub Free allows unlimited private repository, this is my default hosting service for Git.
Data Processing
Conservation palaeobiology is a data driven discipline. At some point in your workflow, you will need to read your data and analyze it. In academia, the programming language explicitly developed to do this is R, which is a free software environment for statistical computing and graphics. The R community is very friendly and will help you throughout your learning experience. Another open source programming language with a wider-scope (a so called general-purpose language) is Python. Python is easier to maintain and more robust than R and further provides cutting-edge access to machine learning algorithms. A third language in data science on the rise is Julia, which beats R in compilation speed. I have used both R and Python in the past and would recommend that beginners start with R and then learn Python later on. Julia is currently too young to provide robust code packages but will be a very good alternative in the future.
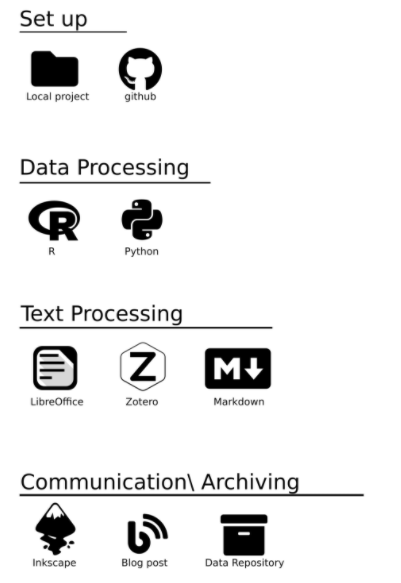
Text Processing
The data is analyzed and the results are promising. Now you need to write down your findings. Microsoft Word is a commonly used tool but is a proprietary software. A free and open source alternative is LibreOffice. A big advantage is that it will work on Linux distributions as well (in contrast to Word). To keep track of sources and references, I have made some very good experiences with Zotero. It is currently my default tool to collect, organize, cite, and share research. It further works well with Markdown, a plain markup language that I use when I want to host my text on the web (such as this blog-post). Note that an alternative to LibreOffice is LaTex. I really like the scope behind LaTex but never used it as I normally collaborate with many coworkers on a text, and not all of them will be able to work with LaTex (while LibreOffice documents can be transformed to word format).
Communication / Archiving
Communication of your results to the wider public commonly works through an article. All submission systems allow LibreOffice files. You can simply reference your code using GitHub during your submission to further show the robustness of your analysis (see my blog-post on how to do this for single- or double-blind peer reviews). Any figures or plots can be modified with Inkscape, a vector graphics software. Note that you should only add minor modification to your figures using Inkscape, but figures should be generally produced using programming code to ensure reproducibility. For the same scope, you should host your code and data in a public repository. An alternative to an article is hosting your results on the internet. There are a lot of open source tools to do this, but I get good results with Hugo combined with Netlify. My personal website, for example, was completely built using open source tools including R, Markdown, and Hugo.
Conclusion
These tools will normally be sufficient for a robust workflow in science. They will facilitate your work and ensure that people can reproduce any results, as well as that the code will still work in a few years. This is especially important for conservation palaeobiologists, where the robustness of results are very important for both stake-holders and decision-makers.

Author: Gregor Mathes
Gregor is a PhD student in Analytical Paleontology at the University of Bayreuth

